Parallel Computing (CMSC416/CMSC616)
Assignment 2: MPI
Due: October 9 10, 2024 @ 11:59 PM Eastern Time
The purpose of this programming assignment is to gain experience in parallel programming on a cluster and MPI. For this assignment, you have to write a parallel implementation of a program to simulate the Game of Life.
Serial Algorithm 1
The game of life simulates simple cellular automata. The game is played on a rectangular board containing cells. At the start, some of the cells are occupied, the rest are empty. The game consists of constructing successive generations of the board. The rules for constructing the next generation from the previous one are:
- death: cells with 0,1,4,5,6,7, or 8 neighbors die (0,1 of loneliness and 4-8 of over population)
- survival: cells with 2 or 3 neighbors survive to the next generation.
- birth: an unoccupied cell with 3 neighbors becomes occupied in the next generation.
You can use the provided serial code as a baseline to develop your parallel implementation. It provides some basic functionality such as parsing the input file and exporting the final board state to a CSV file. Your task is to implement the parallel version using C, C++, or Fortran, and MPI. You can adapt this Makefile and batch script for your needs.
Input/Initialization
Your program should read in a data file containing the coordinates of cells that are initially alive. Sample files are located here: life.1.256x256.data and life.2.256x256.data (256x256 board). Each line in this file represents the coordinates of a cell on the board that is live. For instance, the following entry:
1,3
Your program should take four command line arguments: the name of the data file, the number of generations to run before stopping the program, X_limit, and Y_limit (representing the size of the board). To be more specific, the command line of your program should be:
./life <data-file-name> <# of generations> <X_limit> <Y_limit>
The number of processes used to launch the program is specified as part of the
mpirun command with the -np argument.
mpirun -np <# of processes> ./life <data-file-name> <# of generations> <X_limit> <Y_limit>
Output
Your program should write a single file called
<data-file-name>.<no-of-generations>.csv (from one
designated rank) that contains comma separated values representing the coordinates of the live cells on the board.
There should be one line (containing the x coordinate, a comma, and then the y
coordinate) for each occupied cell at the end of the last generation/iteration.
Sample output files are available:
- life.1.256x256.100.csv is the output of the file life.1.256x256.data run for 100 generations on a 256x256 board
- life.2.256x256.100.csv is the output of the file life.2.256x256.data run for 100 generations on a 256x256 board
The only print from your program to standard output should be from process 0 that looks like this:
TIME: Min: 25.389 s Avg: 27.452 s Max: 41.672 s
// C++:
cout << "TIME: Min: " << local_time << " s Avg: " << local_time << " s Max: " << local_time << " s\n";
// C:
printf("TIME: Min: %f s Avg: %f s Max: %f s\n", local_time, local_time, local_time);
Parallel Version
Figure out how you will decompose the problem for parallel execution. Remember that MPI (at least the OpenMPI implementation) does not always have great communication performance and so you will want to make message passing infrequent. Also, you will need to be concerned about static load balancing during data distribution/domain decomposition. You should use the 1D decomposition (over rows) and implement it using non-blocking Isend/Irecv calls.
You can assume that X_limit and Y_limit will be powers of 2 as will be the number of processes you will be running on. You can also assume that you will be running the program on a minimum of 4 processes and X_limit and Y_limit are much larger than the number of processes.
When you need to distribute the initial state of the board to different processes, you can read the entire file on one process (say rank 0) and then send messages from rank 0 to everyone else.
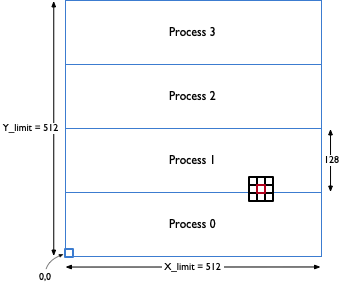
- Keeping a copy of the entire board on each process and/or using collectives to send ghost rows between processes is the WRONG method for parallelizing this problem.
- If the data you are trying to send is not contiguous in memory, you need to first copy it to a contiguous buffer and then point to that buffer in the MPI_Isend call. Similar considerations for MPI_Irecv.
What to Submit
You must submit the following files and no other files:
life-nonblocking.[c,cpp,f77,f90]: parallel version using non-blocking Isend/Irecv routines, where the file extension depends on the language used for the implementationMakefilethat will compile your code successfully on zaratan when using mpicc or mpicxx, and generate this executable:life-nonblocking.- You must also submit a short PDF report (called
report-assign2.pdf) with performance results (a line plot). The line plot should present the execution times to run the parallel code on the input file life.512x512.data (for 4, 8, 16, 32, 64, and 128 processes), running on a 512x512 board for 500 iterations. In total, you will be running the program 6 times. In the report, you should mention:- how was the initial data distribution done
- what are the performance results, and are they what you expected
LastName-FirstName-assign2), compress it to .tar.gz
(LastName-FirstName-assign2.tar.gz) and upload that to gradescope.- Make sure that the generated executable name is life-nonblocking, and do not include the executable in the tarball.
- Use MPI_Wtime to time the code and MPI reduction to compute min, max, and avg runtime. Only time the main compute loop (not including the time for distributing the input board, gathering the final state, etc).
- Use the compiler flag -g while debugging but -O2 when collecting performance numbers for the report.
- Make sure that your batch script has the --exclusive flag when collecting execution times.
Tips
- Zaratan primer
- MPI_Wtime example
Grading
The project will be graded as follows:
| Component | Percentage |
|---|---|
| Runs correctly with 4 processes | 30 |
| Runs correctly with 16 processes | 40 |
| Performance with 4 processes | 10 |
| Performance with 16 processes | 10 |
| Writeup | 10 |
We will compare your performance numbers with our parallel solution as the baseline. You will get full points as long as your performance is within 2 times (2x) of the performance of our solution. For performance worse than 2x of the expected performance, you will receive credit proportional to the ratio of the two.
4 processes: 0.056 s
16 processes: 0.017 s