High Performance Computing (CMSC714)
Project 1: MPI
Due: September 28, 2023 @ 6:00 PM
The purpose of this programming project is to gain experience in parallel programming on a cluster and MPI. For this project you have to write a parallel implementation of an iterative stencil computation.
We will use a two-dimensional (2D) Jacobi calculation to simulate heat diffusion. This calculation performs iterative sweeps (called timesteps) over a given array of double precision floating point values (so C or C++ double types) . The serial algorithm is described here and you have to implement its parallel version using C or C++, and MPI.
Input/Initialization
Your program should read in a file containing the values that will be used to initialize the 2D array of doubles. A sample file is available here. This sample input corresponds to the initial conditions in the figure to the right. Two regions of size 128x128 are assigned initial values of 212 and 32 degrees F (Fahreneheit) respectively to denote hot and cold zones. All remaining array elements are assigned an initial temperature of 72 degrees F.
Your program should take four command line arguments: the name of the data file, the number of iterations or timesteps, and the X and Y sizes of the 2D array, size_X, and size_Y. To be more specific, the command line of your program should be: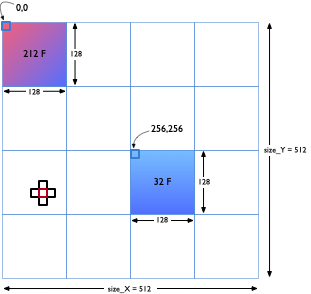
./jacobi2d <input filename> <# of timesteps> <size_X> <size_Y>
For the sample file above, size_X = size_Y = 512. The number of processes the program will run on is specified as part of the mpirun command with the -np argument.
mpirun -np <# of processes> ./jacobi2d <input filename> <# of timesteps> <size_X> <size_Y>
Serial Computation
At each timestep, the value in each cell (array element) is updated by averaging the values of its four neighbors and itself.
A[i, j] = (A[i, j] + A[i-1, j] + A[i+1, j] + A[i, j-1] + A[i, j+1]) * 0.2
You can assume periodic boundary conditions so the array elements at the edges will exchange data with elements on the opposite edge.
Output
Your program should write a single file called <size_X>x<size_Y>.<#-processes>-output.csv (from one designated rank) that contains comma separated values (up to three decimal points) of the 2D array. Each line in the file should correspond to one row of the array starting with i=0, j=0.
This is the correct output file for the sample input file above after 100 timesteps.
The only print from your program to standard output should be from process 0 that looks like this:
TIME: Min: 25.389 s Avg: 27.452 s Max: 41.672 s
Parallel Version
Use a 2D decomposition (both rows and columns) to parallelize the program. You can assume that size_X and size_Y will be powers of 2 as will be the number of processes you will be running on. You can also assume that you will be running the program on a minimum of 4 processes and size_X and size_Y are much larger than the number of processes.
Other sample inputs and outputs after 100 timesteps:
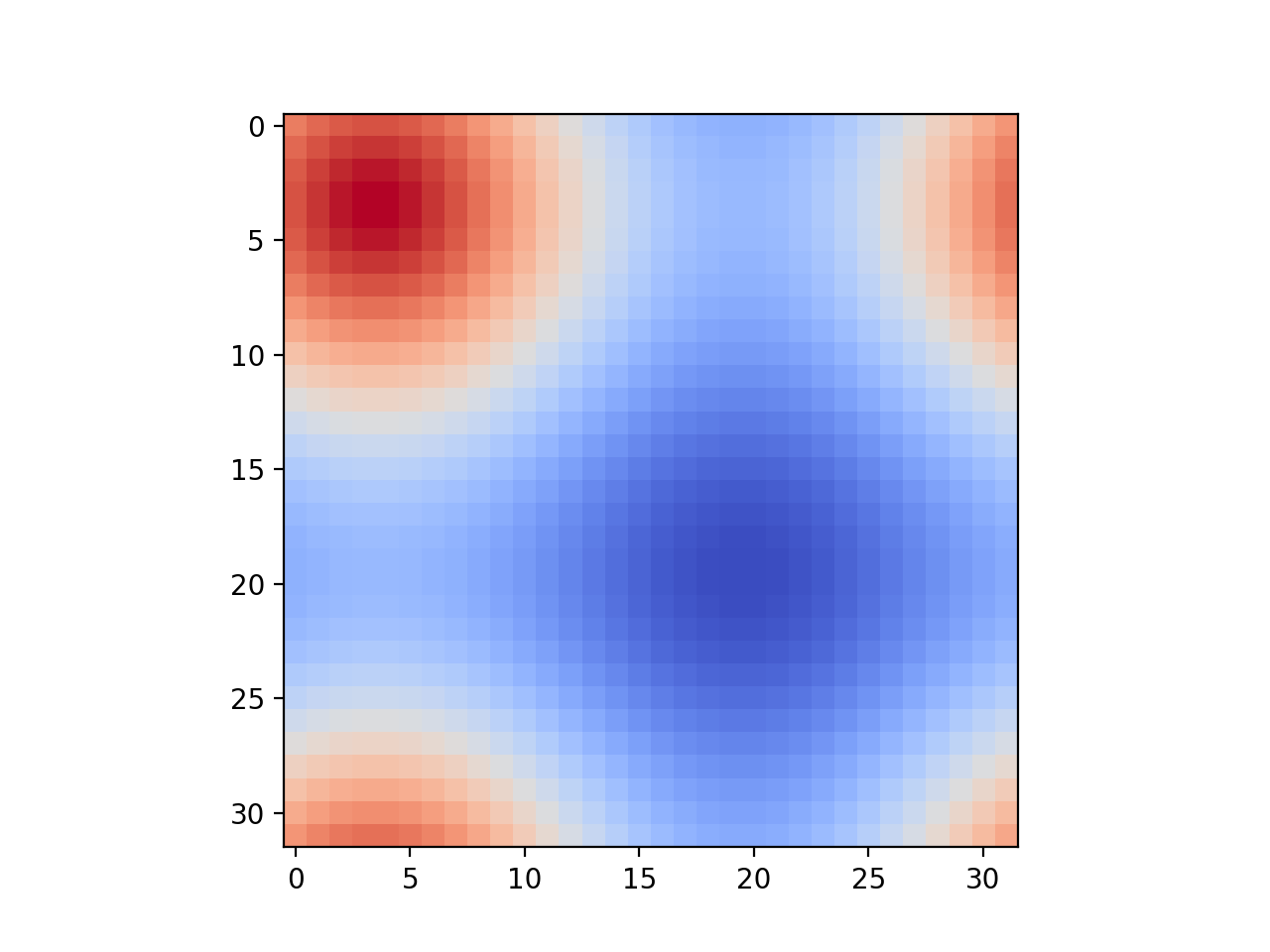
- Input 1 Output 1 size_X = 32, size_Y = 32, timesteps = 100
- Input 2 Output 2 size_X = 64, size_Y = 128, timesteps = 100
- Input 3 Output 3 size_X = 128, size_Y = 256, timesteps = 100
- Input 4 Output 4 size_X = 1024, size_Y = 1024, timesteps = 500
What to Submit
You must submit the following files and no other files:
jacobi2d.[c,C]: your parallel implementationMakefilethat will compile your code successfully on zaratan when using mpicc or mpicxx. You can see a sample Makefile here. Make sure that the executable name is jacobi2d and do not include the executable in the tarball.- You must also submit a short report (pdf) with performance results (a line plot). The line plot should present the execution times to run the parallel version on the sample 1024x1024 input file (for 4, 8, 16, 32, and 64 processes) for 500 iterations. Since the cluster you run on has 128 cores per node, you will only run on 1 node so will not be using the Infiniband network.
LastName-assign1), compress it to .tar.gz (LastName-assign1.tar.gz) and upload that to ELMS.
Tips
- Quick zaratan primer.
- Use -g while debugging but -O2 when collecting performance numbers.
- MPI_Wtime example
- You can use this Python script to visualize the output csv at any timestep.
Grading
The project will be graded as follows:
| Component | Percentage |
|---|---|
| Runs correctly on 4 processes | 10 |
| Runs correctly on 64 processes | 40 |
| Performance using 4 processes | 20 |
| Speedup with 64 processes | 20 |
| Writeup | 10 |